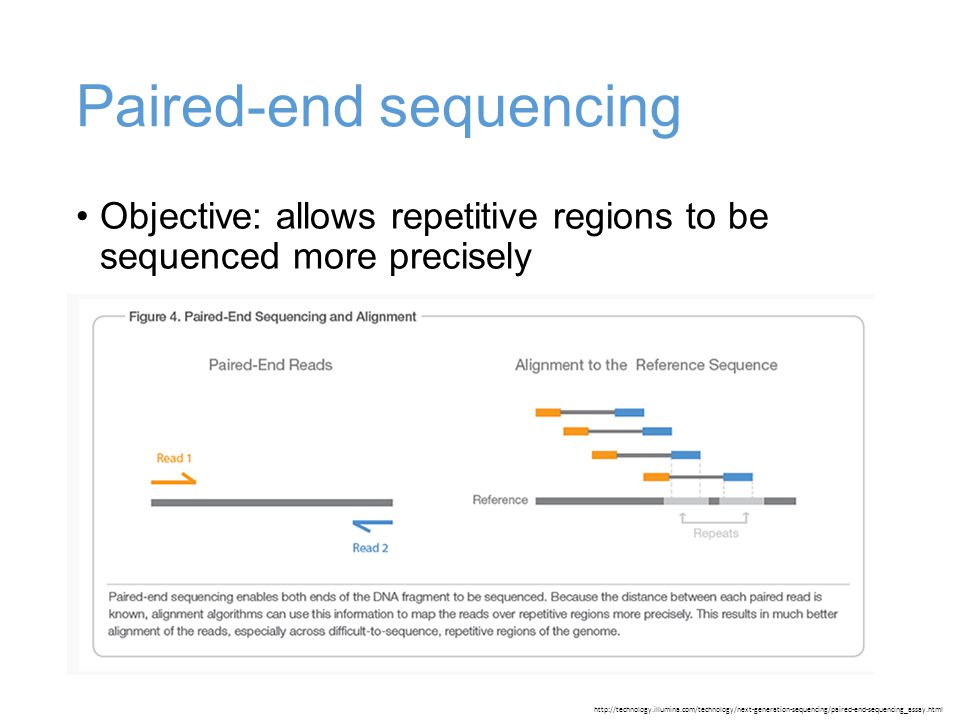
paired end sequencing vs mate pair
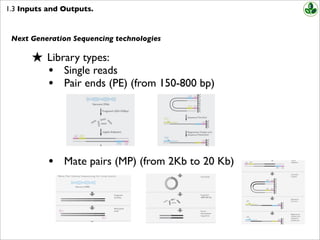
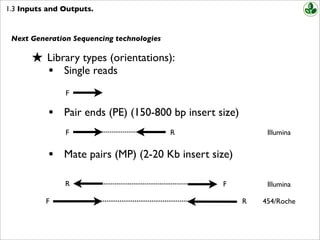
Combining data generated from mate pair library sequencing with that from short-insert paired-end reads provides a powerful combination of read lengths for maximal sequencing coverage across the genome. Mate pair protocols add to the utility of paired-end sequencing by boosting the genomic distance spanned by each pair of reads potentially allowing larger repeats to be bridged and resolved.
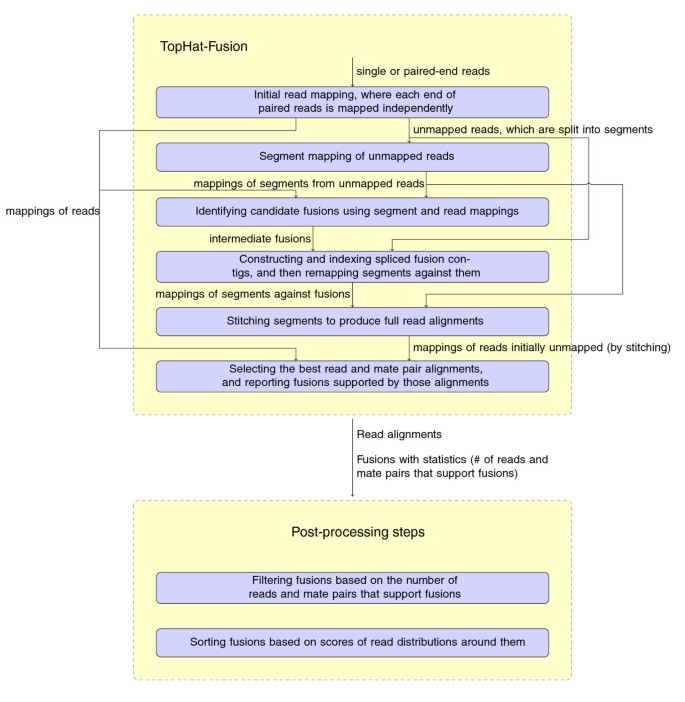
Tophat Fusion An Algorithm For Discovery Of Novel Fusion Transcripts Springerlink
Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data.
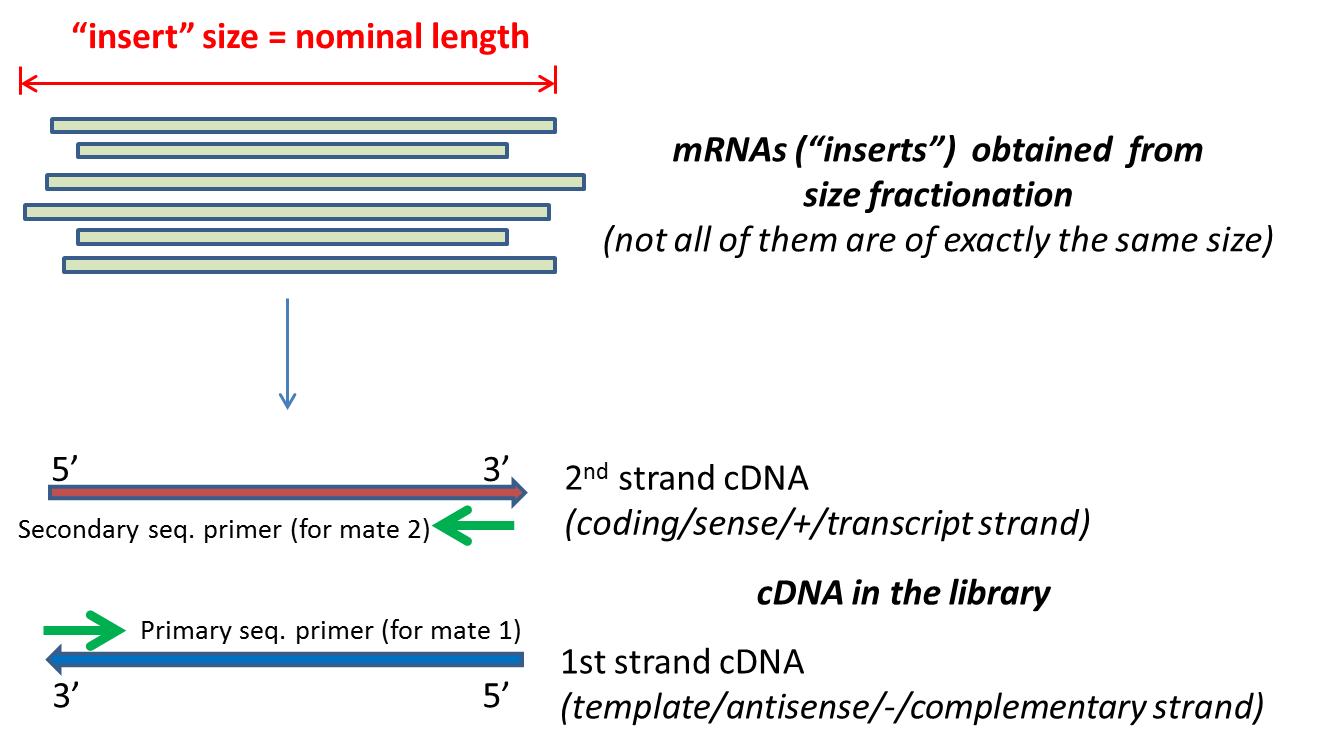
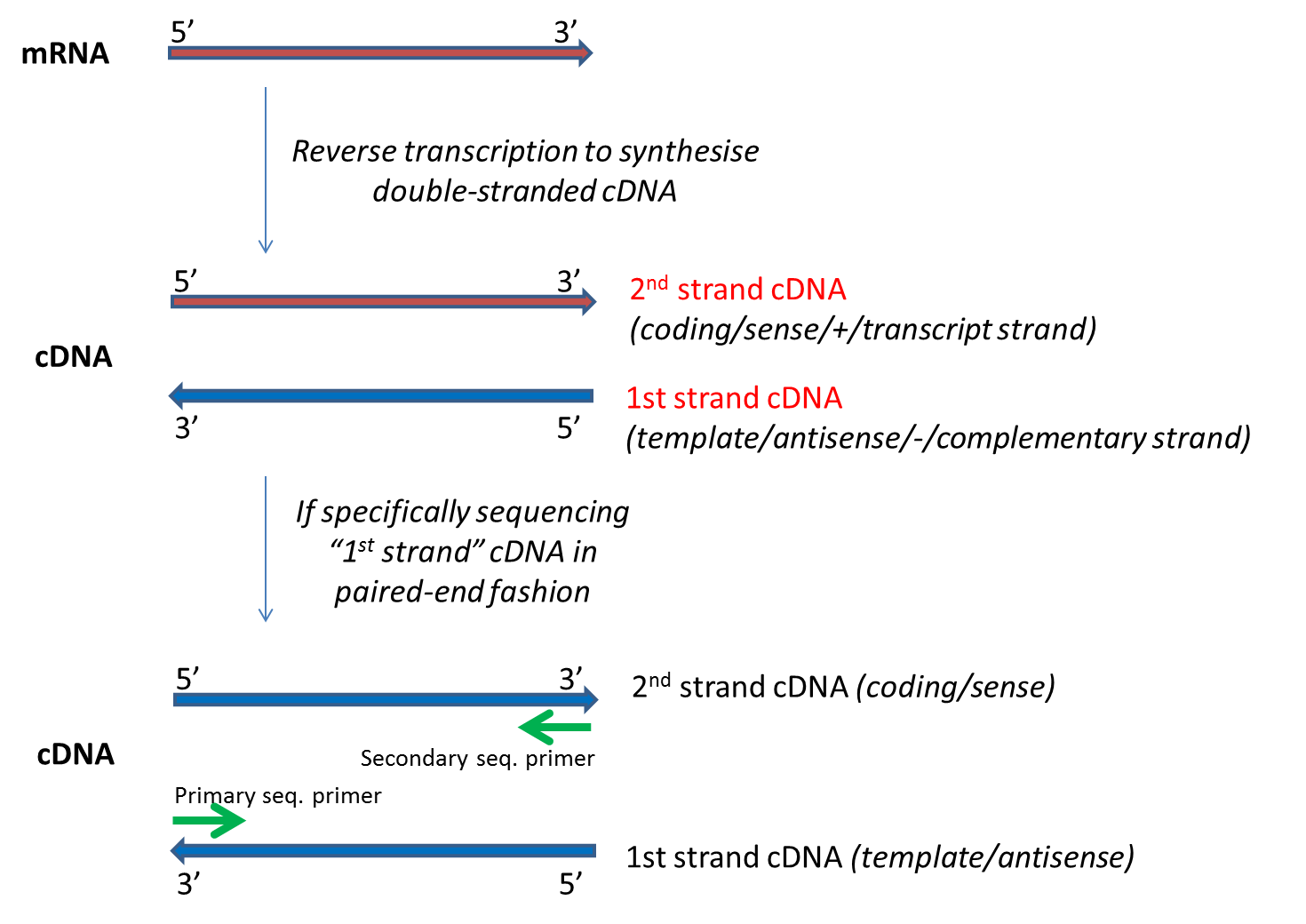
. Since the beginning of 2013 this preparation has been based on Nextera technology. Illumina has this to say on the subject. Illumina gets sequence data from both strands of input sequence which means it outputs data from both ends of the input and is normally reported two files R1 and R2 often refereed to as mates files R1first mates R2second mates.
If --discard-untrimmed is used this means that the entire pair is discarded if R1 or R2 are untrimmed. Mate pair sequencing involves generating long-insert paired-end DNA libraries useful for a number of sequencing applications including. Methods A novel algorithm that utilizes a suffix array has been specifically designed to compute the uniqueness of paired reads with fixed or variable mate-pair distance.
Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts. Combining data generated from mate pair library sequencing with that from short-insert paired-end reads provides a powerful combination of read lengths for maximal sequencing coverage across the genome.
Scaffolds NNN Contigs Illumina Reads Assembly Strategy. The diagrams show the paired-end reads R1 R2 derived from sequencing DNA fragments white boxes with sequencing adapters gray boxes on either end. For example if you have a 300bp contiguous fragment the machine will sequence eg.
Paired-end DNA sequencing also detects common DNA rearrangements such as insertions deletions and inversions. Mate pair sequencing involves generating long-insert paired-end DNA libraries useful for a number of sequencing applications including. Paired-end tags exist in PET libraries with the intervening DNA absent that is a PET represents a larger fragment of.
In addition to producing twice the number of reads for the same time and effort in library. In mate-pair sequencing the library preparation yields two. The preparation of mate pair libraries is designed to allow.
1 shows a schematic view of an Illumina paired-end read. Learn about the difference between Paired-End and Single-Run sequencing and why the former creates more precise alignments than the latter especiall. Read 1 often called the forward read extends from the Read 1 Adapter in the 5 3 direction towards Read 2 along the forward DNA strand.
There is a unique adapter sequence on both ends of the paired-end read labeled Read 1 Adapter and Read 2 Adapter. Popular Answers 1 7th Aug 2017. In revenge for the long-reads I imagine that they are simply reads that are synthesized with a large read size but that do not allow like the maite-pair.
Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. Paired-end sequencing facilitates detection of genomic. For classical paired-end.
Paired-end DNA sequencing reads provide high-quality alignment across DNA regions containing repetitive sequences and produce long contigs for de novo sequencing by filling gaps in the consensus sequence. Since paired-end reads are more likely to align to a reference the quality of the entire data set. Paired-end sequencing facilitates detection of genomic.
The Illumina Nextera Mate Pair NMP protocol uses a circularization-based strategy that leaves behind 38-bp adapter sequences which must be computationally removed. Paired-end tags are the short sequences at the 5 and 3 ends of a DNA fragment which are unique enough that they exist together only once in a genome therefore making the sequence of the DNA in between them available upon search or upon further sequencing. Due to the way data is reported in these files special care has to be taken.
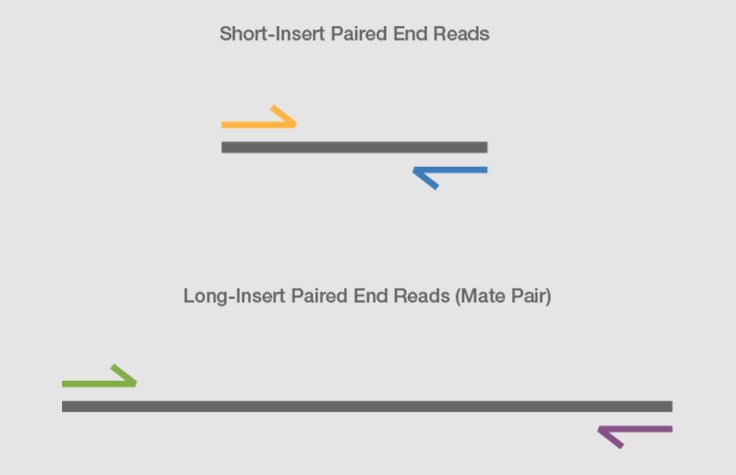
Paired end や mate pair という用語はどのようにライブラリが作られたかどうやってシーケンスされたかを示します どちらの手法も配列情報に加えてゲノム中における二つの read 間の物理的な距離の情報を与えます. We obtain theoretical read length lower bounds for re-sequencing that are also applicable to paired-end de novo assembly. The insert size on classic paired-end is smaller about 500bp while the insert size of mate-pair is much longer several Kb which allows to join the contiguous between them especially is it.
The figure shows the workflow for mate-pair library preparation for Illumina sequencing. Bases 1-75 forward direction and bases 225-300 reverse direction of the fragment. In paired-end sequencing the library preparation yields a set of fragments and the machine sequences each fragment from both ends.
Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts. What and when use Single vs paired end sequences in RNA sequence. The preparation of mate pair libraries is designed to allow classical paired-end sequencing of both ends of a fragment with an original size of several kilobases.
Illumina Paired End Sequencing.

Construction Of The Interval Adjacency Graph Construction Of The Download Scientific Diagram
The Insert Size In Paired End Data Seqanswers

Integrative Genomics Viewer Igv Visualization Of L V Panamensis Download Scientific Diagram

Lapparecchio Catrame Mai Reads Illumina Portare Stagno Capriola
Lapparecchio Catrame Mai Reads Illumina Portare Stagno Capriola
Lapparecchio Catrame Mai Reads Illumina Portare Stagno Capriola

July 2015 Bioinformatics Bits And Pieces

Figure 1 From Paired De Bruijn Graphs A Novel Approach For Incorporating Mate Pair Information Into Genome Assemblers Semantic Scholar

Pdf A Field Guide To Whole Genome Sequencing Assembly And Annotation

Mapping Structural Variants Using The Paired End Read Strategy A A Download Scientific Diagram

Preparation Of Dna Sequencing Libraries For Illumina Systems 6 Key Steps In The Workflow Thermo Fisher Scientific Fr

Mate Pair Sequencing Improves Detection Of Genomic Abnormalities In Acute Myeloid Leukemia Aypar 2019 European Journal Of Haematology Wiley Online Library